Менделевская рандомизация с двумя выборками: практика написания кода, изучите ее за один шаг
Загрузите и установите пакет R.
#install.packages("rlang")
library(rlang)
# Install packages
#install.packages("remotes")
#remotes::install_github("MRCIEU/TwoSampleMR")
#remotes::install_github("MRCIEU/ieugwasr")
#remotes::install_github("MRCIEU/MRInstruments")
#remotes::install_github("n-mounier/MRlap")
#install.packages("data.table")
#install.packages("devtools")
#install.packages("dplyr")
#install.packages("xlsx")
# Load Packages
library(TwoSampleMR)
library(ieugwasr)
library(MendelianRandomization)
library(MRInstruments)
library(MRlap)
library(dplyr)
library(xlsx)
Получить разоблачение
> gwasinfo("ukb-b-12493") ##Эссенциальная гипертония
id trait
1 ukb-b-12493 Diagnoses - secondary ICD10: I10 Essential (primary) hypertension
note
1 41204#I10: Output from GWAS pipeline using Phesant derived variables from UKBiobank
sample_size build year category ncase consortium ncontrol nsnp
1 463010 HG19/GRCh37 2018 Binary 54358 MRC-IEU 408652 9851867
group_name population priority author ontology unit sex mr
1 public European 1 Ben Elsworth NA SD Males and Females 1
subcategory
1 NA
hyperten_tophits <- tophits(id="ukb-b-12493", clump=0)
hyperten_gwas <- rename(hyperten_tophits, c(
"SNP"="rsid",
"effect_allele.exposure"="ea",
"other_allele.exposure"="nea",
"beta.exposure"="beta",
"se.exposure"="se",
"eaf.exposure"="eaf",
"pval.exposure"="p",
"N"="n"))
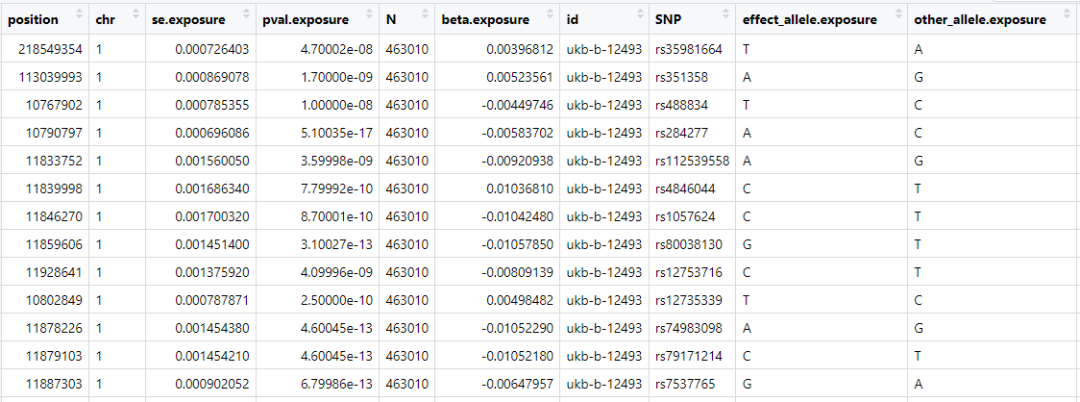
hyperten_liberal <- clump_data(hyperten_gwas, clump_kb = 10000, clump_r2 = 0.05,) ##Параметры регулируются
## Removing 2685 of 2773 variants due to LD with other variants or absence from LD reference panel
hyperten_liberal$id.exposure = "hyperten - liberal clump"
Вычислить значение F и значение R^2
# Calculate R2 and F stat for exposure data
# Liberal hypertension F stat
hyperten_liberal$r2 <- (2 * (hyperten_liberal$beta.exposure^2) * hyperten_liberal$eaf.exposure * (1 - hyperten_liberal$eaf.exposure)) /
(2 * (hyperten_liberal$beta.exposure^2) * hyperten_liberal$eaf.exposure * (1 - hyperten_liberal$eaf.exposure) +
2 * hyperten_liberal$N * hyperten_liberal$eaf.exposure * (1 - hyperten_liberal$eaf.exposure) * hyperten_liberal$se.exposure^2)
hyperten_liberal$F <- hyperten_liberal$r2 * (hyperten_liberal$N - 2) / (1 - hyperten_liberal$r2)
hyperten_liberal_meanF <- mean(hyperten_liberal$F)
Получить информацию, соответствующую результатам, основанным на воздействии.
# Find SNPs in outcome data
ao <- available_outcomes()
anxiety_hyperten_liberal <- extract_outcome_data(snps = hyperten_liberal$SNP, outcomes = "ukb-b-11311")
##
Extracting data for 88 SNP(s) from 1 GWAS(s)
Finding proxies for 34 SNPs in outcome ukb-b-11311
Extracting data for 34 SNP(s) from 1 GWAS(s)
Server code: 502; Server is possibly experiencing traffic, trying again...
Server code: 502; Server is possibly experiencing traffic, trying again...
Server code: 502; Server is possibly experiencing traffic, trying again...
Retry succeeded!
Вы также можете проверить финальную информацию——
gwasinfo("ukb-b-11311") ##беспокойство
id trait
1 ukb-b-11311 Diagnoses - secondary ICD10: F41.9 Anxiety disorder, unspecified
note
1 41204#F419: Output from GWAS pipeline using Phesant derived variables from UKBiobank
sample_size build year category ncase consortium ncontrol nsnp
1 463010 HG19/GRCh37 2018 Binary 1523 MRC-IEU 461487 9851867
group_name population priority author ontology unit sex mr
1 public European 1 Ben Elsworth NA SD Males and Females 1
subcategory
1 NA
Люблю слышать и видеть гармонию
# Harmonise datasets
harm_anxiety_hyperten_liberal <- harmonise_data(exposure_dat = hyperten_liberal, outcome_dat = anxiety_hyperten_liberal)
##
Harmonising hyperten (hyperten - liberal clump) and Diagnoses - secondary ICD10: F41.9 Anxiety disorder, unspecified || id:ukb-b-11311 (ukb-b-11311)
Removing the following SNPs for being palindromic with intermediate allele frequencies:
rs1870735, rs35184780
> dim(harm_anxiety_hyperten_liberal)
[1] 55 43
МР-анализ
anxiety_hyperten_liberal_results <- mr(dat = harm_anxiety_hyperten_liberal)

Обратите внимание на количество nsnp в данный момент ~ Почему не хватает двух?
# F stats for each analysis
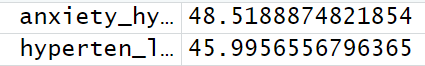
anxiety_hyperten_liberal_meanF <- mean(harm_anxiety_hyperten_liberal$F)
Сравните два F-средних и посмотрите, есть ли какие-нибудь изменения~
После достижения этой точки,На самом деле это хорошо сочетается с твитами прошлой недели.➡TwoSampleMR: комплексный анализ менделевской рандомизацииПодключено,Все, попробуйте

Неразрушающее увеличение изображений одним щелчком мыши, чтобы сделать их более четкими артефактами искусственного интеллекта, включая руководства по установке и использованию.
Копикодер: этот инструмент отлично работает с Cursor, Bolt и V0! Предоставьте более качественные подсказки для разработки интерфейса (создание навигационного веб-сайта с использованием искусственного интеллекта).
Новый бесплатный RooCline превосходит Cline v3.1? ! Быстрее, умнее и лучше вилка Cline! (Независимое программирование AI, порог 0)

Разработав более 10 проектов с помощью Cursor, я собрал 10 примеров и 60 подсказок.
Я потратил 72 часа на изучение курсорных агентов, и вот неоспоримые факты, которыми я должен поделиться!
Идеальная интеграция Cursor и DeepSeek API
DeepSeek V3 снижает затраты на обучение больших моделей
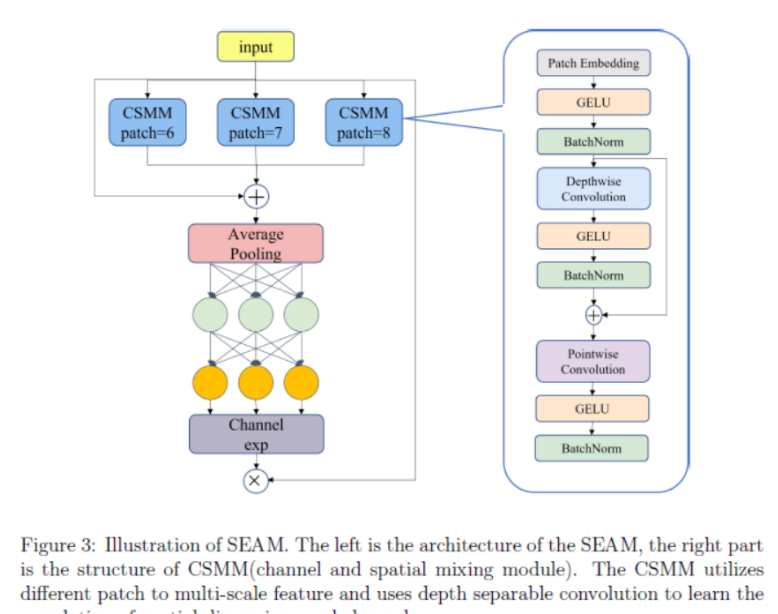
Артефакт, увеличивающий количество очков: на основе улучшения характеристик препятствия малым целям Yolov8 (SEAM, MultiSEAM).

DeepSeek V3 раскручивался уже три дня. Сегодня я попробовал самопровозглашенную модель «ChatGPT».

Open Devin — инженер-программист искусственного интеллекта с открытым исходным кодом, который меньше программирует и больше создает.
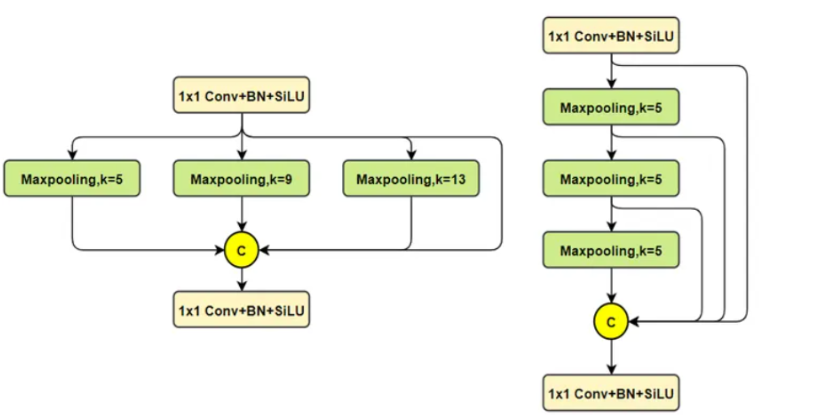
Эксклюзивное оригинальное улучшение YOLOv8: собственная разработка SPPF | SPPF сочетается с воспринимаемой большой сверткой ядра UniRepLK, а свертка с большим ядром + без расширения улучшает восприимчивое поле

Популярное и подробное объяснение DeepSeek-V3: от его появления до преимуществ и сравнения с GPT-4o.

9 основных словесных инструкций по доработке академических работ с помощью ChatGPT, эффективных и практичных, которые стоит собрать
Вызовите deepseek в vscode для реализации программирования с помощью искусственного интеллекта.
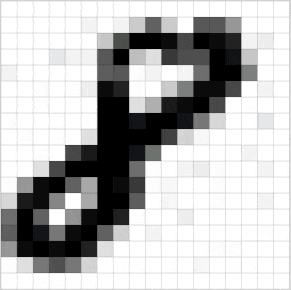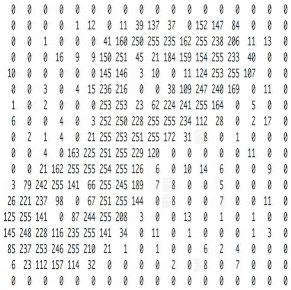
Познакомьтесь с принципами сверточных нейронных сетей (CNN) в одной статье (суперподробно)
50,3 тыс. звезд! Immich: автономное решение для резервного копирования фотографий и видео, которое экономит деньги и избавляет от беспокойства.
Cloud Native|Практика: установка Dashbaord для K8s, графика неплохая
Краткий обзор статьи — использование синтетических данных при обучении больших моделей и оптимизации производительности
MiniPerplx: новая поисковая система искусственного интеллекта с открытым исходным кодом, спонсируемая xAI и Vercel.
Конструкция сервиса Synology Drive сочетает проникновение в интрасеть и синхронизацию папок заметок Obsidian в облаке.
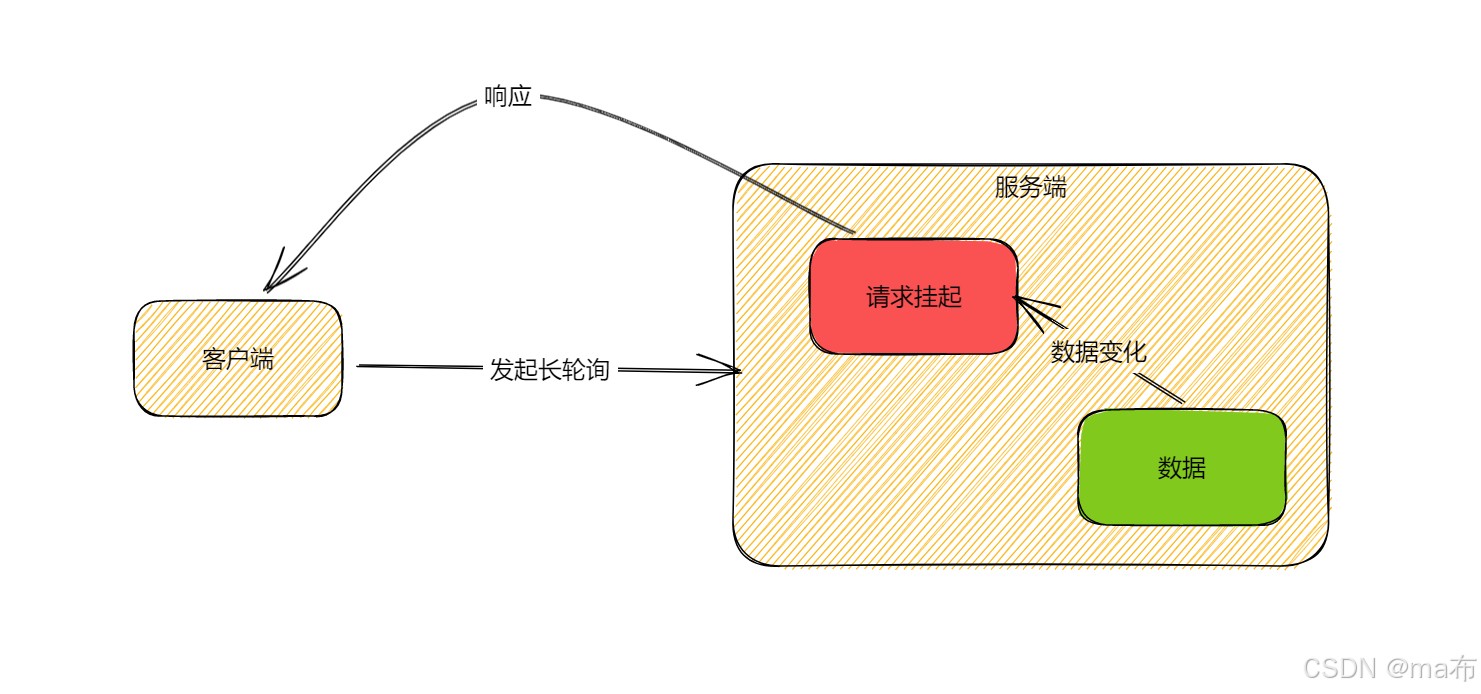
Центр конфигурации————Накос
Начинаем с нуля при разработке в облаке Copilot: начать разработку с минимальным использованием кода стало проще

[Серия Docker] Docker создает мультиплатформенные образы: практика архитектуры Arm64

Обновление новых возможностей coze | Я использовал coze для создания апплета помощника по исправлению домашних заданий по математике

Советы по развертыванию Nginx: практическое создание статических веб-сайтов на облачных серверах
Feiniu fnos использует Docker для развертывания личного блокнота Notepad

Сверточная нейронная сеть VGG реализует классификацию изображений Cifar10 — практический опыт Pytorch
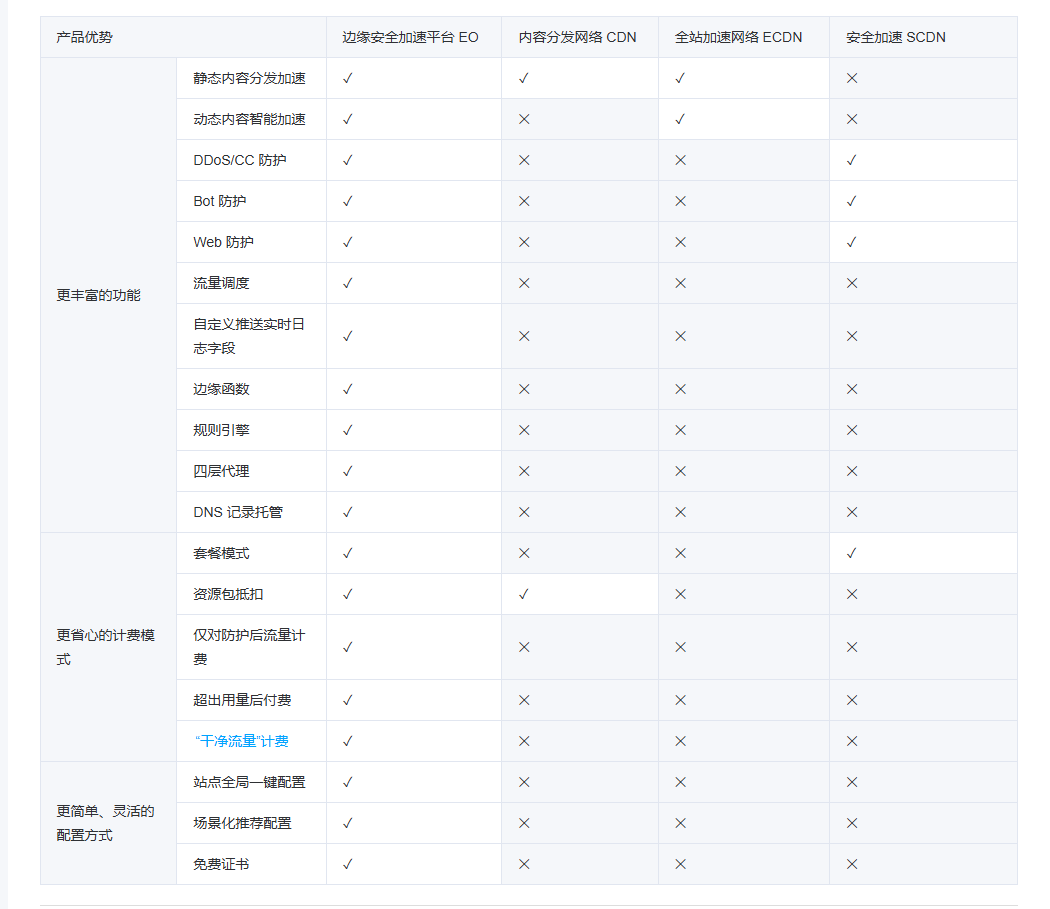
Начало работы с EdgeonePages — новым недорогим решением для хостинга веб-сайтов
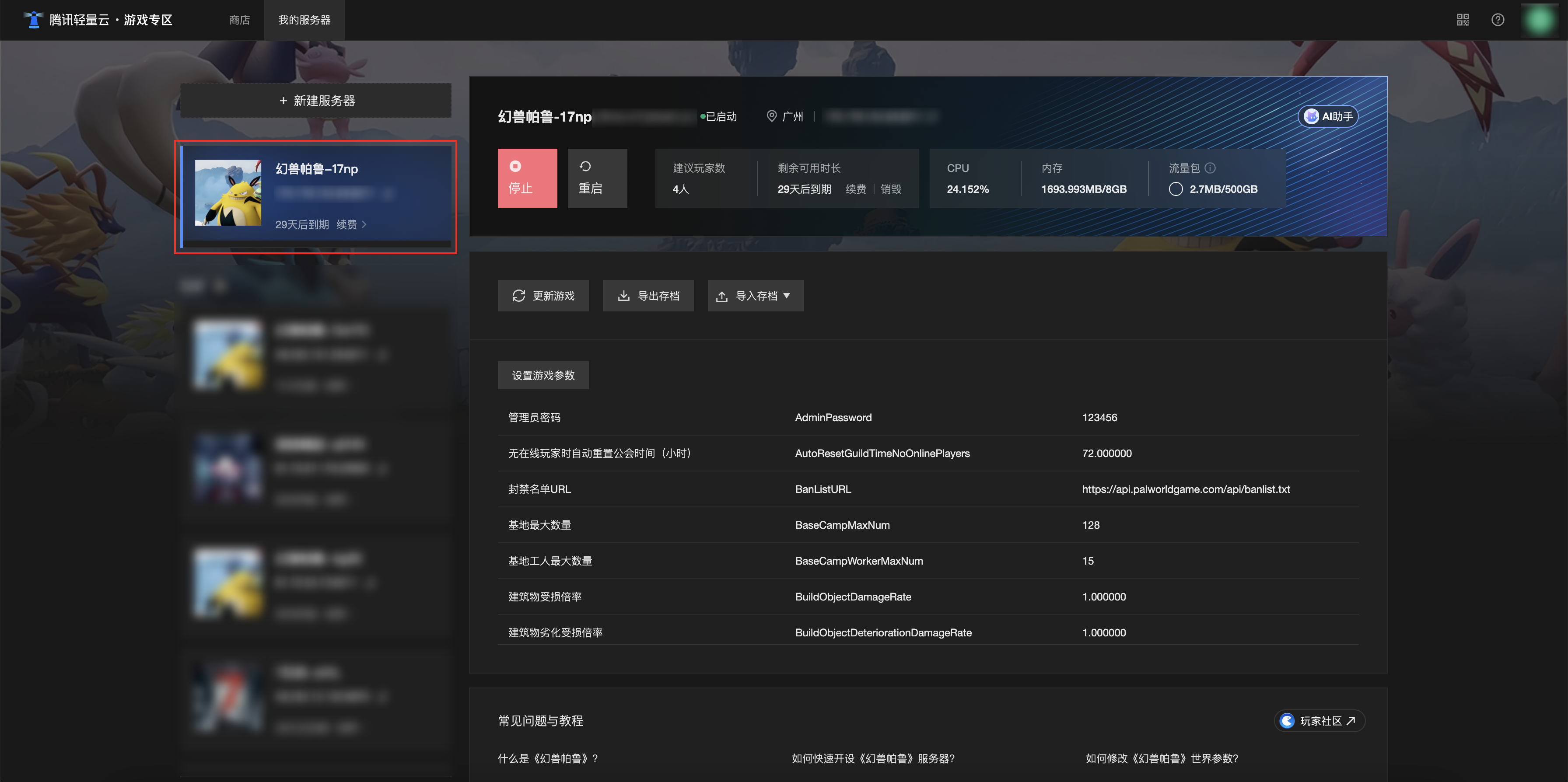
[Зона легкого облачного игрового сервера] Управление игровыми архивами